clip seq紫外交联的作用
CLIP-seq,又称为HITS-CLIP,即紫外交联免疫沉淀结合高通量测序(crosslinking-immunoprecipitation and high-throughput sequencing), 是一项在全基因组水平揭示RNA分子与RNA结合蛋白相互作用的革命性技术。
其主要原理是基于RNA分子与RNA结合蛋白在紫外照射下发生耦联,以RNA结合蛋白的特异性抗体将RNA-蛋白质复合体沉淀之后,回收其中的RNA片段,经添加接头、RT-PCR等步骤,对这些分子进行高通量测序,再经生物信息学的分析和处理、总结,挖掘出其特定规律,从而深入揭示RNA结合蛋白与RNA分子的调控作用及其对生命的意义。
Cross-Linking and Immunoprecipitation (CLIP)
Cross-Linking and Immunoprecipitation (CLIP) is an antibody-based technique used to study RNA-protein interactions related to RNA immunoprecipitation (RIP), but differs from RIP in the use of UV radiation to cross-link RNA and the binding proteins. Unlike DNA-protein crosslinking which is done with formaldehyde, CLIP uses ultraviolet (UV) light and the crosslinking is irreversible. UV crosslinks are also more specific and only link proteins to RNAs that are in very close proximity. Further, UV crosslinks do not form between two proteins. For these reasons, UV has become the standard in RNA research. The RNA-RBP complexes are then immunoprecipitated. Due to the irreversibility of the crosslinks, the next step is digestion with a proteinase.
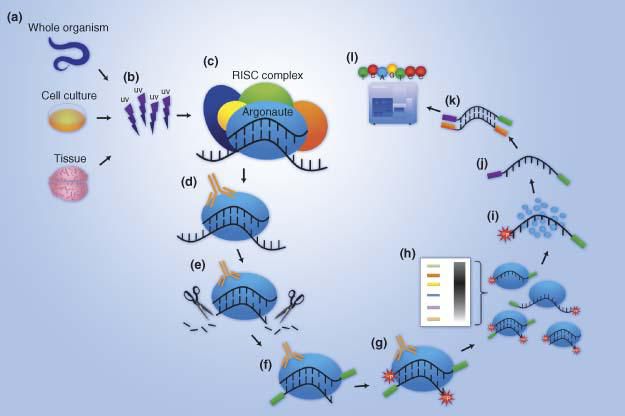
UV crosslinking immunoprecipitation (CLIP) is an increasingly popular technique to study protein-RNA interactions in tissues and cells. Whole cells or tissues are ultraviolet irradiated to generate a covalent bond between RNA and proteins that are in close contact. After partial RNase digestion, antibodies specific to an RNA binding protein (RBP) or a protein-epitope tag is then used to immunoprecipitate the protein-RNA complexes. After stringent washing and gel separation the RBP-RNA complex is excised. The RBP is protease digested to allow purification of the bound RNA. Reverse transcription of the RNA followed by high-throughput sequencing of the cDNA library is now often used to identify protein bound RNA on a genome-wide scale. UV irradiation can result in cDNA truncations and/or mutations at the crosslink sites, which complicates the alignment of the sequencing library to the reference genome and the identification of the crosslinking sites. Meanwhile, one or more amino acids of a crosslinked RBP can remain attached to its bound RNA due to incomplete digestion of the protein. As a result, reverse transcriptase may not read through the crosslink sites, and produce cDNA ending at the crosslinked nucleotide. This is harnessed by one variant of CLIP methods to identify crosslinking sites at a nucleotide resolution. This method, individual nucleotide resolution CLIP (iCLIP) circularizes cDNA to capture the truncated cDNA and also increases the efficiency of ligating sequencing adapters to the library. Here, we describe the detailed procedure of iCLIP.








 添加微信咨询!
添加微信咨询!